mafa
mafa.RmdCompute orientations
Load data of parcels of the city of Rouen in 1827 and extract segments (lines) from polygons.
# transform polygons as segments (lines)
linerouen <- mafa_geom_to_segment(sfobject = rouen_1827, to = 'LINESTRING')morphal_geom_to_segment() can be applied on LINESTRING,
e.g. in the case of streets modeled as lines.
Compute orientations of segments with East looking and perpendicular parameters.
orientationsest <- mafa_segment_orientation(sfsegments = linerouen, looking = 'E', perpendicular = TRUE)
orientationsest |>
ggplot() +
geom_sf(aes(color = orientation)) +
scale_color_viridis_c() +
theme_bw() +
theme(axis.ticks = element_blank(), axis.text = element_blank(), panel.grid = element_blank())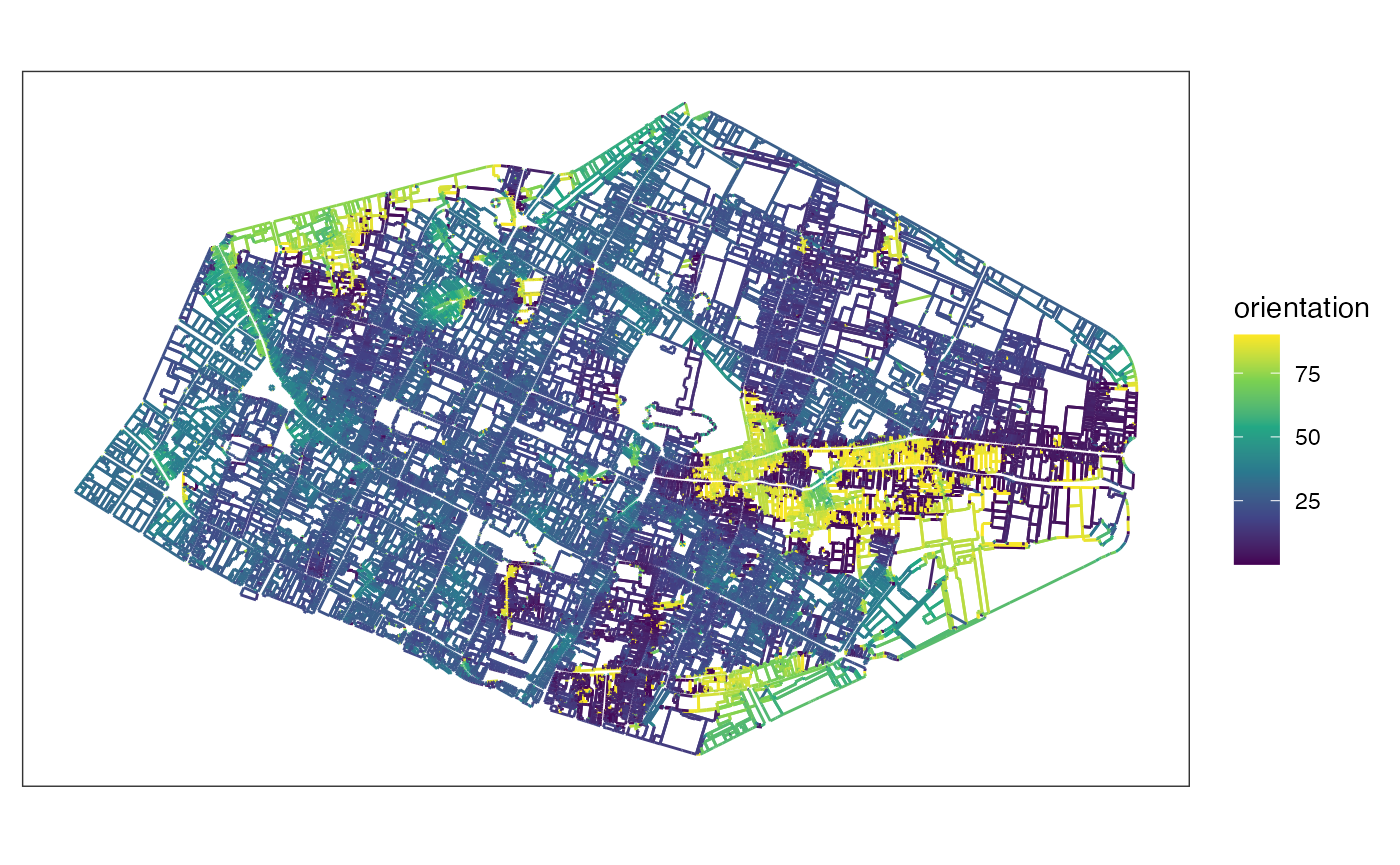
Compute morphological indices of polygons
Example with distance to minimal bounding rectangle (DSR):
rouen_1827 |>
mafa_dsr() |>
ggplot() +
geom_sf(aes(fill = dsr), color = 'grey90', linewidth = 0.05) +
scale_fill_viridis_c(direction = -1) +
theme_bw() +
theme(axis.ticks = element_blank(), axis.text = element_blank(), panel.grid = element_blank())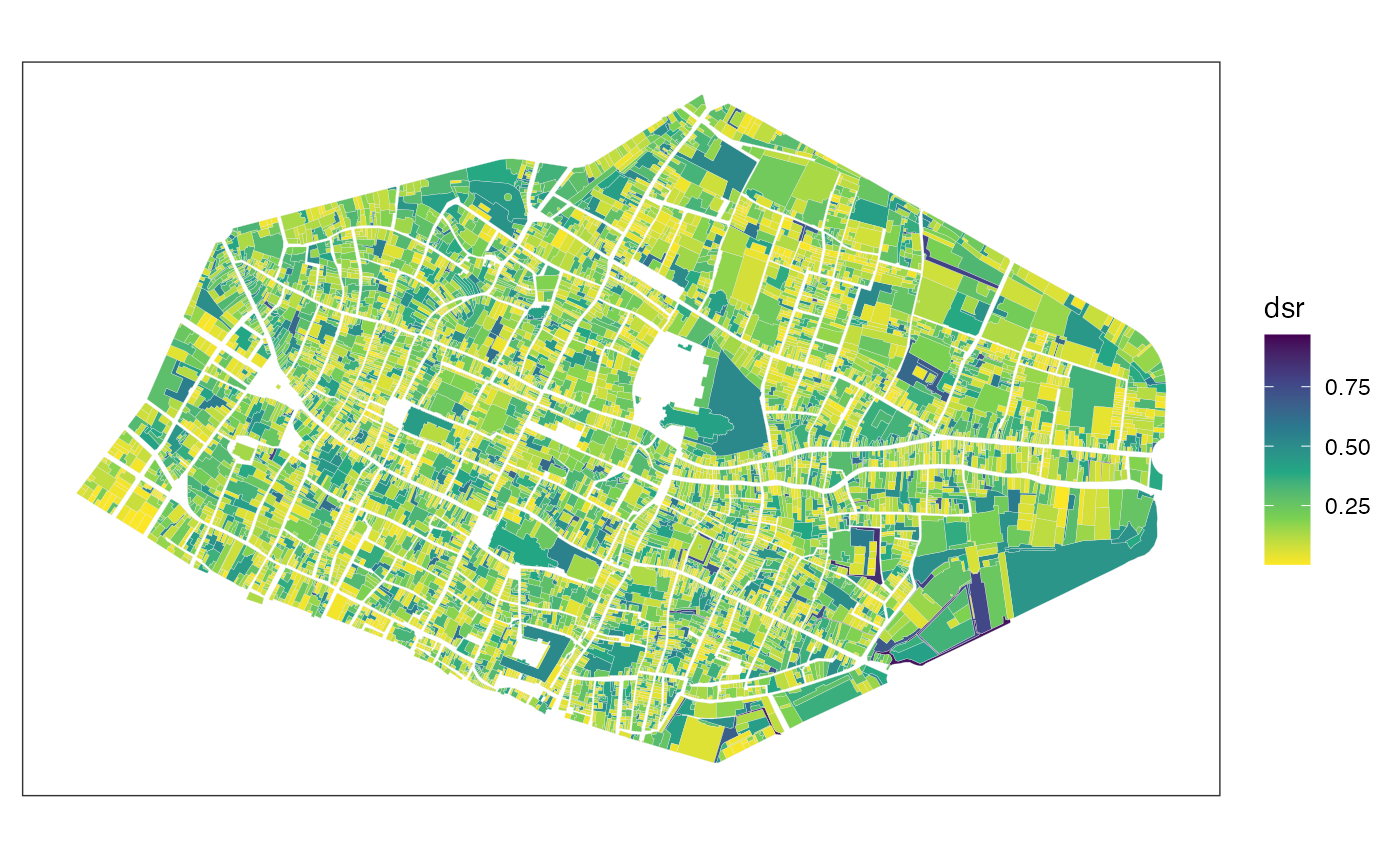
Compute clusters from indices
Compute indices.
rouen_with_indices <- rouen_1827 |>
mafa_circularity() |>
mafa_dsc() |>
mafa_dsr()
rouen_with_indices <- rouen_with_indices[,2:5] # without rowid
rouen_with_indices
#> Simple feature collection with 10240 features and 3 fields
#> Geometry type: POLYGON
#> Dimension: XY
#> Bounding box: xmin: 560973.9 ymin: 6927999 xmax: 563141.3 ymax: 6929327
#> Projected CRS: RGF93 v1 / Lambert-93
#> # A tibble: 10,240 × 4
#> geometry miller_index dsc dsr
#> <POLYGON [m]> <dbl> <dbl> <dbl>
#> 1 ((561754.2 6928994, 561759.5 6928992, 561756.2… 0.690 0 0.174
#> 2 ((561779.4 6929002, 561773.6 6928982, 561764.7… 0.689 -3.19e-12 0.0625
#> 3 ((561732.2 6928970, 561735.3 6928967, 561741.4… 0.247 2.30e- 1 0.405
#> 4 ((561715.6 6928974, 561708.7 6928977, 561705.1… 0.180 5.55e- 1 0.717
#> 5 ((561605.9 6928902, 561603.4 6928896, 561592.8… 0.719 0 0.0265
#> 6 ((561604.7 6928915, 561607.3 6928914, 561614 6… 0.490 3.56e- 3 0.0603
#> 7 ((561610.9 6928904, 561611.7 6928904, 561610 6… 0.493 2.79e- 4 0.0623
#> 8 ((561621 6928889, 561612.5 6928867, 561608.2 6… 0.441 1.20e- 2 0.0711
#> 9 ((561627.2 6928869, 561623 6928871, 561619.3 6… 0.773 6.85e- 3 0.0271
#> 10 ((561628.3 6928906, 561621.3 6928890, 561616.4… 0.680 2.91e- 2 0.122
#> # ℹ 10,230 more rowsCreate HCA clustering and plot result:
clusterrouen <- mafa_clustering(sf = rouen_with_indices, pca_center = TRUE, pca_scale = TRUE, hca_method = 'ward.D2')
plot(clusterrouen, labels = FALSE, hang = 0)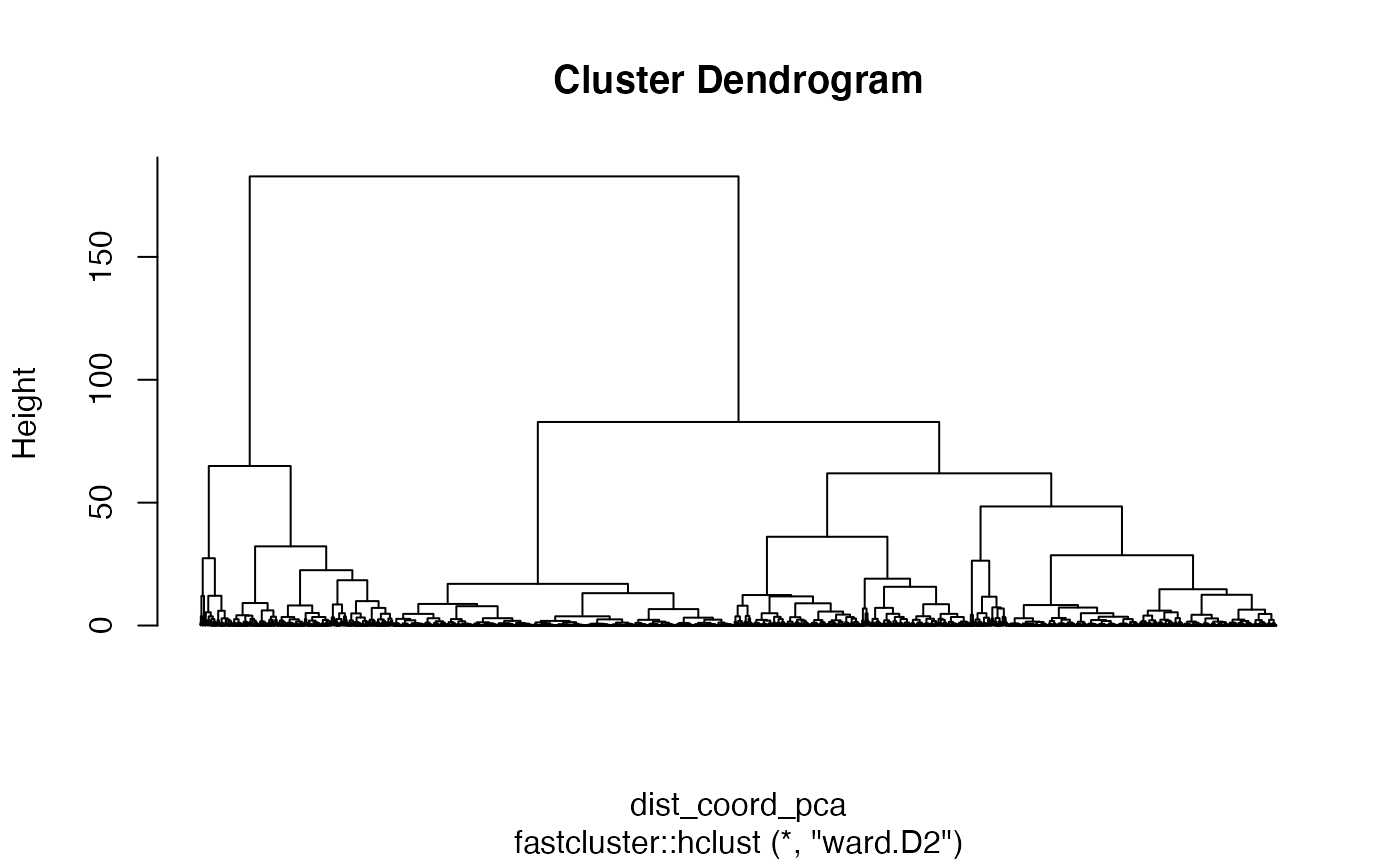
Clusters
Cartography of clusters:
mafa_clusters(sf = rouen_with_indices, clustering = clusterrouen, cutting = 6) |>
ggplot() +
geom_sf(aes(fill = cluster), color = 'white', linewidth = 0.02) +
ggthemes::scale_fill_colorblind() +
theme_bw() +
theme(axis.ticks = element_blank(), axis.text = element_blank(), panel.grid = element_blank())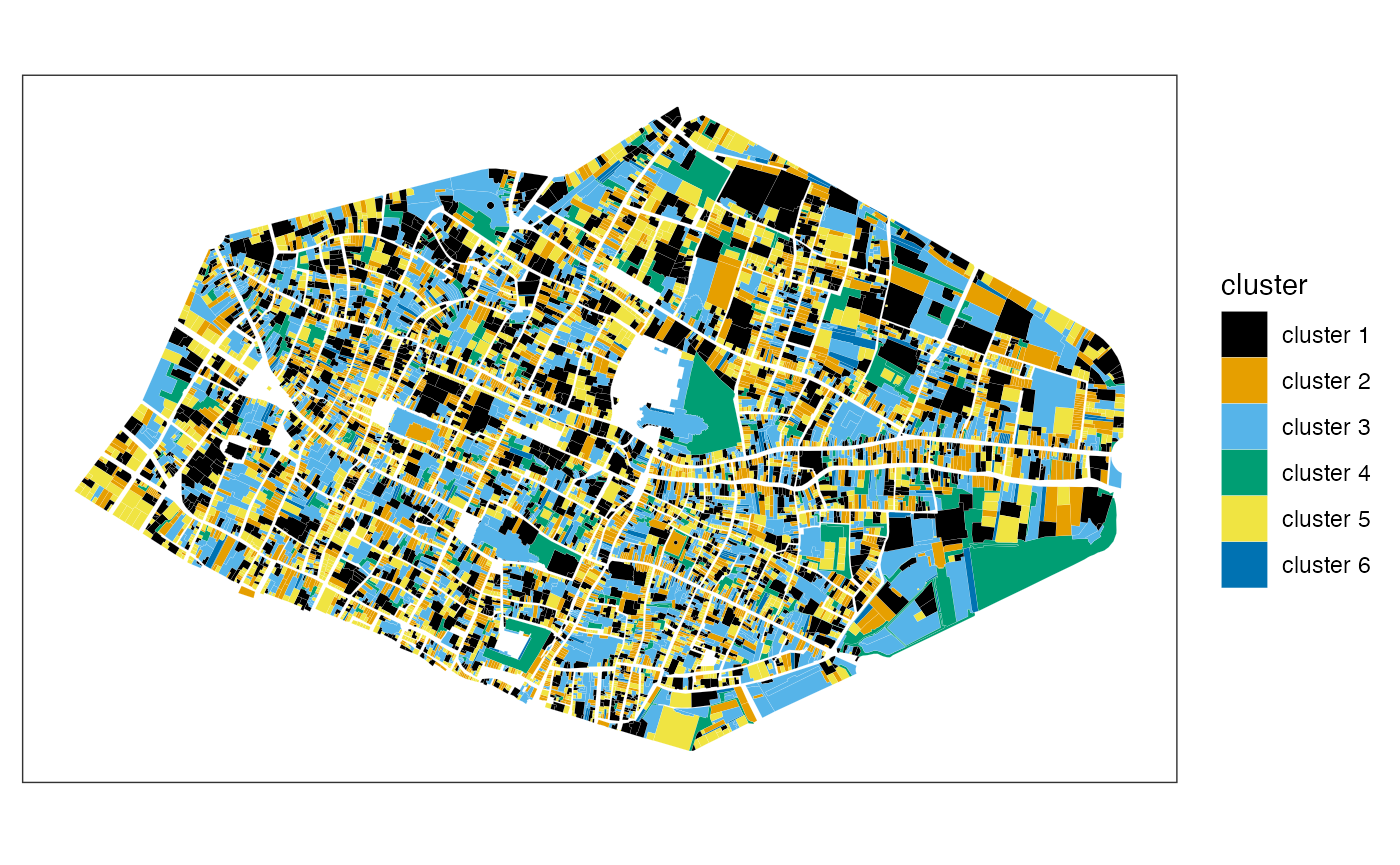
Summary of clusters with mean of center-scale values of variables by cluster.
mafa_clusters_mean(sf = rouen_with_indices, clustering = clusterrouen, cutting = 6)
#> # A tibble: 6 × 4
#> cluster miller_index dsc dsr
#> <fct> <dbl> <dbl> <dbl>
#> 1 cluster 1 0.131 0.118 0.367
#> 2 cluster 2 -0.152 -0.551 -0.632
#> 3 cluster 3 -1.14 1.38 1.38
#> 4 cluster 4 -2.20 3.54 2.92
#> 5 cluster 5 0.991 -0.593 -0.688
#> 6 cluster 6 -1.95 -0.359 0.0322